A recent study published in MBExpress (Springer Nature Group), explored yeast diversity envisaging the efficient utilization of methanol as the sole carbon source for growth.
Methanol is a promising feedstock for biomanufacturing, but the efficiency of methanol-based bioprocesses is limited by the low rate of methanol utilization pathways and methanol toxicity.
Exploring yeast diversity to identify innate robust strains with strong catabolic and biosynthetic capacities and genetic engineering potential, is advantageous for improving bioprocesses efficiency. This study describes the isolation and molecular identification of yeast isolates of the methylotrophic species Candida boidinii from waters derived from the traditional curation of olives, in different years, and from contaminated superficial soil near fuel stations.
The yeast microbiota from these habitats was also characterized. C. boidinii isolates obtained from the curation of olives’ water exhibited significantly higher maximum specific growth rates, compared with the isolates obtained from fuel-contaminated soils, when grown on methanol as the sole C-source (range 0.15–0.19 h−1, compared with 0.05–0.06 h−1, when grown on (1% (v/v), methanol in shake flasks, at 30°C). The isolates exhibit significant robustness towards methanol toxicity that increases as the cultivation temperature decreases from 30°C to 25°C. These methanol-efficient catabolizing isolates are proposed as a promising platform to develop methanol-based bioprocesses.
The full article can be read here https://amb-express.springeropen.com/articles/10.1186/s13568-024-01754-9#ref-CR72
The first author is Marta Mota who developed this research when working for the PhD degree of the PhD program (DP-AEM) in Biotechnology and Biosciences of the Department of Bioengineering, iBB, Instituto Superior Técnico. Margarida Palma contributed to yeast isolation and Professor Isabel Sá-Correia is the corresponding author and the thesis supervisor.
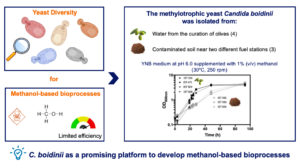 Photo credits: Ganesh Mohan T from Wikimedia Commons under the Licence Creative Commons Attribution-Share Alike 4.0 with no changes.
Photo credits: Ganesh Mohan T from Wikimedia Commons under the Licence Creative Commons Attribution-Share Alike 4.0 with no changes.



